29th October 2017 New and improved version of CRISPR for more precise genetic engineering An improved version of the genetic engineering technique known as CRISPR has been published in the journals Science and Nature.
Two separate studies have demonstrated a powerful new method of genetic engineering, which could be used to improve the research and treatment of diseases in the future. In the first study, scientists directly and permanently changed single base pairs of DNA from A*T to G*C. This could one day enable precise DNA surgery to correct mutations. In the second, RNA was edited rather than DNA, which has potential to treat diseases without permanently affecting the genome. The Howard Hughes Medical Institute, which led the first study, created a new enzyme known as a "base editor". This allows researchers to edit the individual base pairs of DNA that form the instructions of life. By altering the molecular structure of one base – adenine (A), cytosine (C), guanine (G) or thymine (T) – and converting it to another, genetic faults that cause diseases could be fixed with high levels of precision. Researchers can now manipulate all four bases. Howard Hughes scientists took cells from patients and used base editing to correct hemochromatosis, an inherited condition that leads to dangerously high levels of iron in the blood. In DNA, each base on one strand is joined with its "partner" base on an opposing strand – so that, for example, adenine pairs with thymine (A*T), while guanine pairs with cytosine (G*C). Some genome editing tools, such as CRISPR/Cas9, cut both strands of DNA and rely on the cell's own molecular machinery to fill in the gap with a desired DNA sequence. Base editors, however, can rewrite the individual chemical units of DNA. Last year, the team at Howard Hughes described a base editor that could change C*G base pairs into T*A. But they didn't have the ability to convert A*T to G*C, until now. Mutations in which a G*C mutates into an A*T account for nearly half of the roughly 32,000 single point mutations associated with human diseases. "CRISPR is like scissors, and base editors are like pencils," says David Liu, Professor of Chemical Biology. He and his colleagues are now "hard at work trying to translate base editing technology into human therapeutics." The new system – which relies on the evolution of engineered bacterial colonies to generate the enzyme – is a "really exciting addition to the genome engineering toolbox," explains Feng Zhang, molecular biologist at the Broad Institute of MIT and Harvard, who was not involved in this study, but took part in the second. "It's a great example of how we can harness natural enzymes and processes to accelerate scientific research."
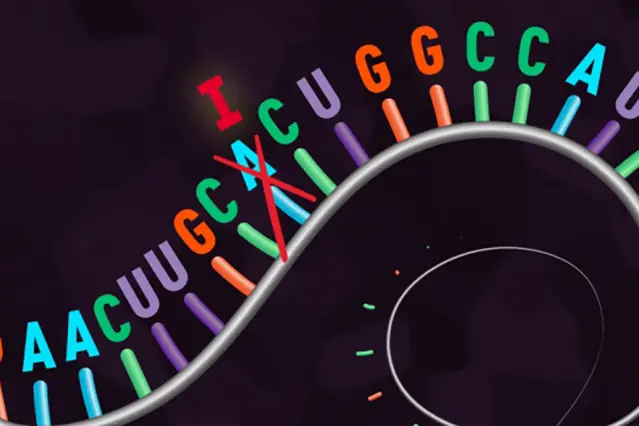
In the second study, by the Broad Institute of MIT and Harvard, researchers adapted CRISPR to edit single RNA letters in human cells. This new system is called RNA Editing for Programmable A to I Replacement, or "REPAIR". RNA is usually single-stranded, and does not form into a double helix as does DNA. Unlike the permanent changes to the genome required for DNA editing, RNA editing provides a safer and more flexible way to make corrections. It has major potential as a tool for disease research and treatment. "REPAIR can fix mutations without tampering with the genome – and because RNA naturally degrades, it's a potentially reversible fix," explains co-author David Cox, a graduate student in Feng Zhang's lab. Like the base editors in the first study, REPAIR has the ability to target individual RNA letters – switching adenosines (A) to inosines (recognised as guanosines (G) by the cell) – without cutting the transcript or relying on the cell's native machinery. These letters are involved in single-base changes, known to regularly cause disease in humans. A mutation from G to A is extremely common; these alterations have been implicated in cases of epilepsy, muscular dystrophy and Parkinson's disease, for example. REPAIR can reverse the impact of any pathogenic G-to-A mutation – regardless of its surrounding letter sequence, with the potential to operate in any cell type. Zhang and his colleagues tested REPAIR on human cells in the laboratory, using this RNA approach to correct an inherited form of anaemia. "The ability to correct disease-causing mutations is one of the primary goals of genome editing," said Professor Zhang. "So far, we've gotten very good at deactivating genes – but actually recovering lost protein function is much more challenging. This new ability to edit RNA opens up more potential opportunities to recover that function and treat many diseases, in almost any kind of cell." Zhang, along with the Broad Institute and MIT, plan to share the REPAIR system widely. As with earlier CRISPR tools, they will make this technology freely available for academic research, via the plasmid-sharing website Addgene, through which the Zhang laboratory has already shared reagents over 42,000 times with researchers at 2,200 labs in 61 countries around the world.
"This is an exciting week for genetic research," said Dr Helen O'Neill, at University College London. "These papers highlight the fast pace of the field and the continuous improvements being made in genome editing, bringing it closer and closer to the clinic." "The science is moving fast in the sense it is becoming less risky, more certain, more precise and more effective," said Dr Sarah Chan, a bioethicist at the University of Edinburgh, in a BBC interview. "It is absolutely past time for us to engage more widely with the public on the issue of gene editing." ---
Comments »
|