9th December 2017 100-fold increase in nanostructure complexity Scientists at the Wyss Institute, part of Harvard University, have announced a 100-fold increase in the complexity of "DNA bricks" that can self-assemble into 3D nanostructures.
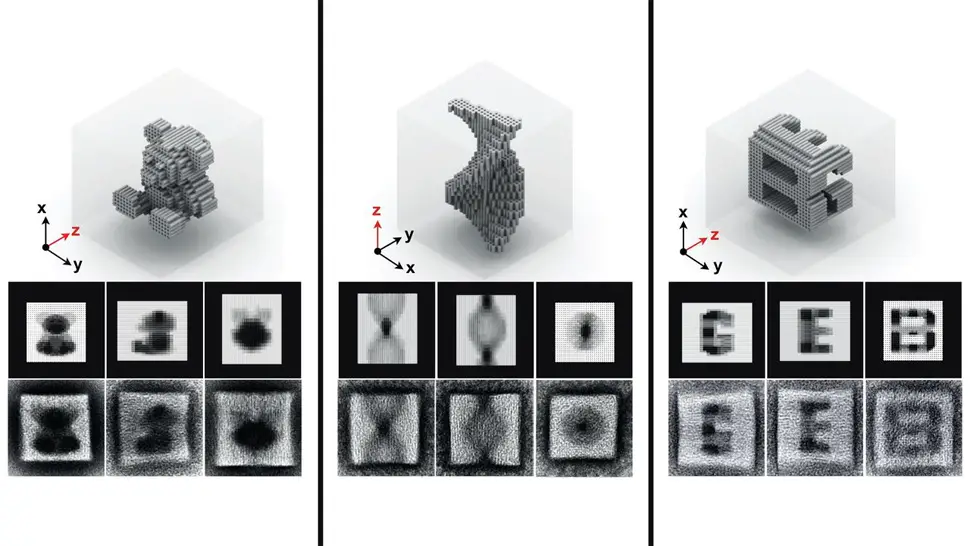
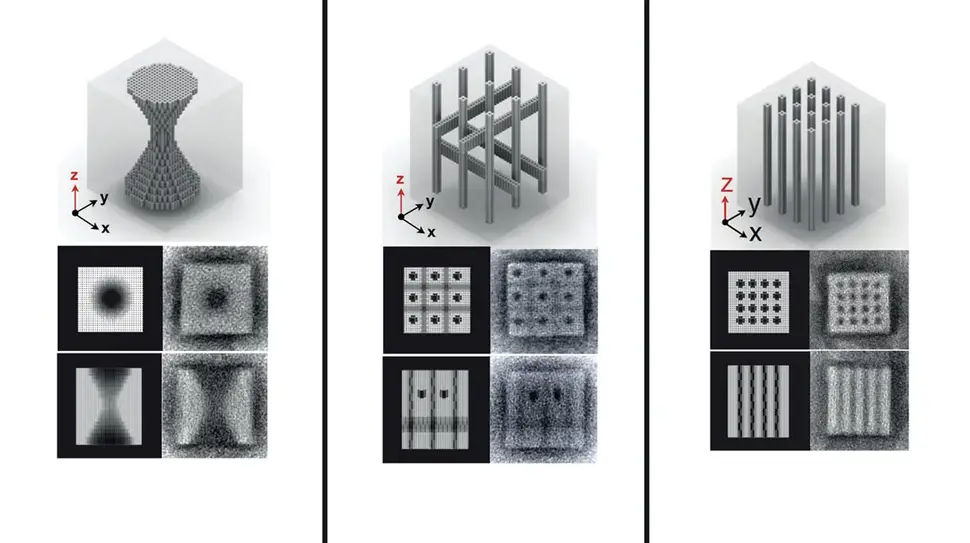
DNA, present in almost every cell, is increasingly being used as a building material to construct tiny, but sophisticated structures – such as smart drug delivery vehicles programmed to release their therapeutic content at disease sites, autonomous 'walkers' that can move along a microparticle surface, fluorescent labels for diagnostic applications, or programmable factories for nanoparticles in optical and electronic applications. To accommodate these functions, researchers at Harvard's Wyss Institute have developed ways that allow DNA strands to self-assemble into increasingly complex 3D structures, such as scaffolded DNA origamis. However, DNA origamis are limited in their sizes, because they rely on the availability of scaffold strands that can be difficult to manufacture and manipulate. In 2012, Peng Yin at Wyss presented an alternative method using DNA 'bricks', which do not use a scaffold, but rather, are able to connect like interlocking Lego bricks and thereby self-assemble into origami-sized structures with prescribed shapes. This month, Yin and his team have leapfrogged their technology by two orders of magnitude, enabling DNA bricks to self-assemble into 3D nanostructures that are 100 times more complex than those made with existing methods. While the first generation DNA bricks could self-assemble from hundreds of components to form nanostructures on the MegaDalton scale, the new bricks allow 10,000 components to form GigaDalton-sized structures (1 GigaDalton = 1000 MegaDaltons or 1 billion Daltons). Yin's method provides user-friendly computational tools for designing DNA nanostructures with complex cavities (and possibly surfaces), with potential to serve as building components in a wide range of nanotechnology applications, from medicine to engineering.
"The principle and promising capabilities of our first-generation DNA bricks led us to ask whether we can enhance the system to attain significantly more complex nanostructures with much higher yields in one-pot assembly reactions. Here we managed to do all this," said Yin, Ph.D., who co-leads the Institute's Molecular Robotics Initiative, and is Professor of Systems Biology at Harvard Medical School. "We worked out an easily accessible practical platform that allows researchers with very different interests and applications in mind to create a molecular canvas with 10,000 bricks, and use it to build nanostructures with unprecedented complexities and potential." "It is remarkable that the bricks were able to distinguish between tens of thousands of potential partners to find their correct neighbours, and it was exciting to see that this technique could be used to form rather complex cavities, such as a teddy bear, the word 'LOVE' or a Möbius strip, amongst many others," comments Luvena Ong, Ph.D., co-author on the study, which is published this month in Nature. In the technology's new version, by varying the length of individual binding domains within the bricks, the team ended up with a substantially increased diversity among possible bricks that, in addition, bind much stronger to each other. "The way the multifaceted DNA bricks technology is evolving shows how the Wyss Institute's Molecular Robotics Initiative can reach deep into the field of DNA nanotechnology to enable new approaches that could solve many real world problems," said Donald Ingber, Ph.D., Wyss Institute Founding Director.
Comments »
If you enjoyed this article, please consider sharing it:
|