DNA storage breakthrough 19th August 2012 For many years, DNA has tantalised scientists with its potential as a storage medium: fantastically dense, stable, energy efficient and proven to work over a timespan of some 3.5 billion years.
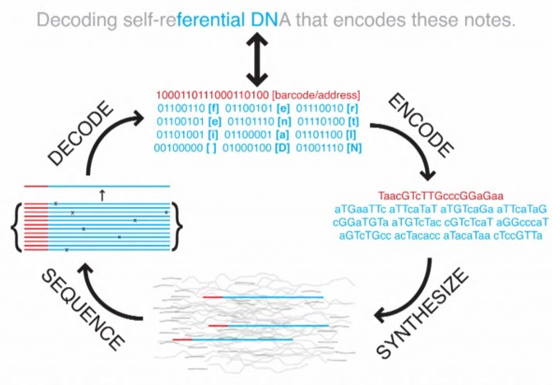
While not the first project to demonstrate the potential of DNA storage, a bioengineer and geneticist at Harvard's Wyss Institute have successfully stored 5.5 petabits of data — around 700 terabytes — in a single gram of DNA, smashing the previous density record by a thousand times. Using next-generation sequencing technology and a novel strategy, Professor George M. Church encoded his book, Regenesis: How Synthetic Biology Will Reinvent Nature and Ourselves, in DNA. It was then copied 70 billion times — roughly triple the sum of the top 100 books of all time — yet small enough to fit on a thumbnail. The team reports its results this week in the journal Science. They used binary code to preserve the text, images and formatting of the book. While the scale is roughly what a 5¼-inch floppy disk once held, the density of the bits is nearly off the charts: 5.5 petabits, or a million gigabits, per cubic millimetre. "The information density and scale compare favorably with other experimental storage methods from biology and physics," said Sri Kosuri, a senior scientist at the institute and senior author on the paper. And where some experimental media — such as quantum holography — require extremely cold temperatures and extremely high energy, DNA is stable at normal room temperature. "You can drop it wherever you want, in the desert or your backyard, and it will be there 400,000 years later," Church said. Reading and writing in DNA is slower than in other media, however, which makes it better suited for archival storage of massive amounts of data, rather than for quick retrieval or data processing. About four grams of DNA could theoretically store the data humankind creates in one year; one bit per base, with each base only a few atoms large.
Although other projects have encoded data in the DNA of living bacteria, the Church team used commercial DNA microchips to create standalone DNA. "We purposefully avoided living cells," Church said. "In an organism, your message is a tiny fraction of the whole cell, so there's a lot of wasted space. But more importantly, almost as soon as a DNA goes into a cell, if that DNA doesn't earn its keep, if it isn't evolutionarily advantageous, the cell will start mutating it, and eventually the cell will completely delete it." In another departure, the team rejected so-called "shotgun sequencing," which reassembles long DNA sequences by identifying overlaps in short strands. Instead, they took their cue from information technology, and encoded the book in 96-bit data blocks, each with a 19-bit address to guide reassembly. Including JPEG images and HTML formatting, the code for the book required 54,898 of these data blocks, each a unique DNA sequence. "We wanted to illustrate how the modern world is really full of zeroes and ones, not As through Zs alone," Kosuri said. The team discussed including a DNA copy with each print edition of Regenesis. But in the book, Church and his co-author, science writer Ed Regis, argue for careful supervision of synthetic biology and policing of its products and tools. Practicing what they preach, the authors decided against an insert — at least until there has been more discussion of the safety, security and ethics of using DNA this way. "Maybe the next book," Church said.
Comments »
|