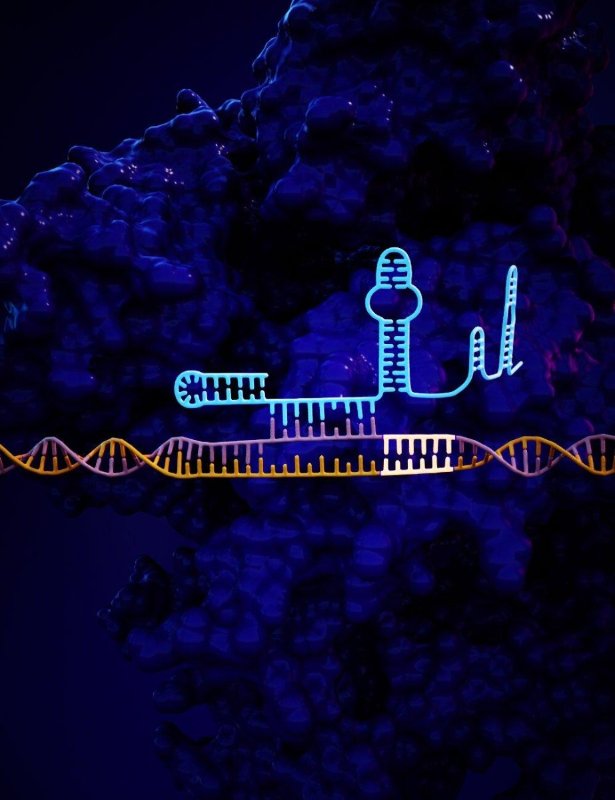
28th April 2019 CRISPR accuracy increased 50-fold Biomedical engineers at Duke University, North Carolina, have developed a method for improving the accuracy of CRISPR genome editing by an average of 50-fold. They believe it can be easily translated to any of the technology's continually expanding formats. The approach adds a short tail to the guide RNA which is used to identify a sequence of DNA for editing. This added tail folds back and binds onto itself, creating a "lock" that can only be undone by the targeted DNA sequence. "CRISPR is generally incredibly accurate, but there are examples that have shown off-target activity, so there's been broad interest across the field in increasing specificity," said Charles Gersbach, Professor of Biomedical Engineering at Duke. "But the solutions proposed thus far cannot be easily translated between different CRISPR systems." "We're focused on a solution that doesn't add more parts and is general to any kind of CRISPR system," said Dewran Kocak, a PhD student working in Gersbach's laboratory who led this project. "What's common to all CRISPR systems is the guide RNA, and these short RNAs are much easier to engineer."
Gersbach and Kocak's solution is to extend the guide RNA by as many as 20 nucleotides in such a way that it folds back onto itself and binds onto the end of the original guide RNA, forming a "hairpin" shape, as seen in the illustration above. This creates a sort of lock that is very difficult to displace if even a single base pair is incorrect in a DNA sequence being scrutinised for a potential cut. But because the guide RNA would prefer to bind to DNA rather than itself, the correct combination of DNA is still able to break the lock. "We're able to fine-tune the strength of the lock just enough so that the guide RNA still works when it meets its correct match," said Kocak. In the paper, Kocak and Gersbach show that this method can increase the accuracy of cuts being made in human cells by an average of 50-fold across five different CRISPR systems derived from four different bacterial strains. And in one case, that improvement rose to over 200-fold. Moving forward, the researchers hope to see just how many different CRISPR variants this approach could work with. And because these experiments were conducted in cultured cells, they are eager to see how well this method can increase CRISPR accuracy within an actual animal model of disease. Their study appears in the journal Nature Biotechnology.
Comments »
If you enjoyed this article, please consider sharing it:
|