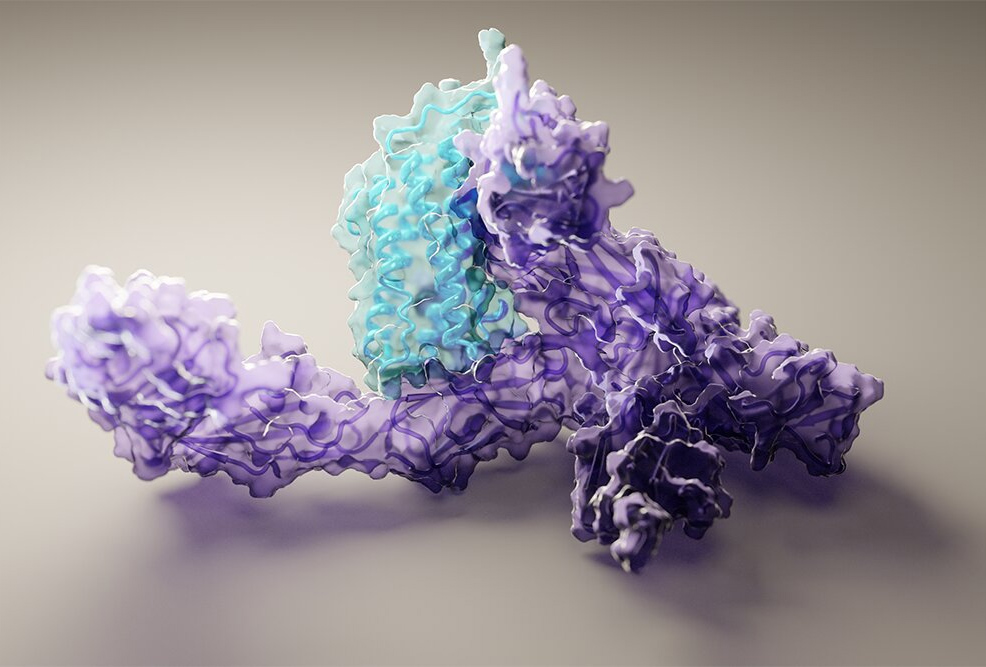
31st July 2021 DeepMind AI predicts 350,000 protein structures Artificial intelligence company DeepMind has mapped the 3D structures of 350,000 proteins, and made the data freely available.
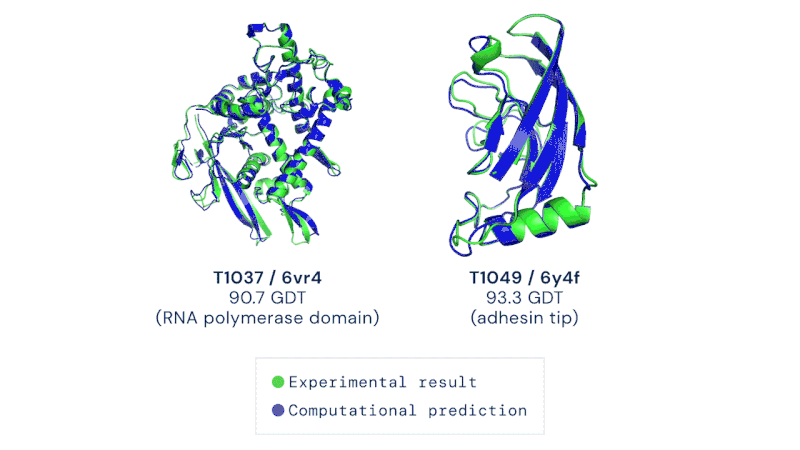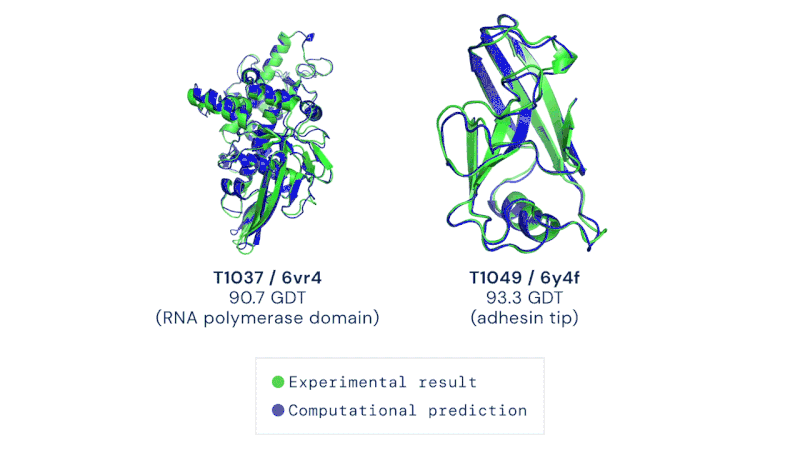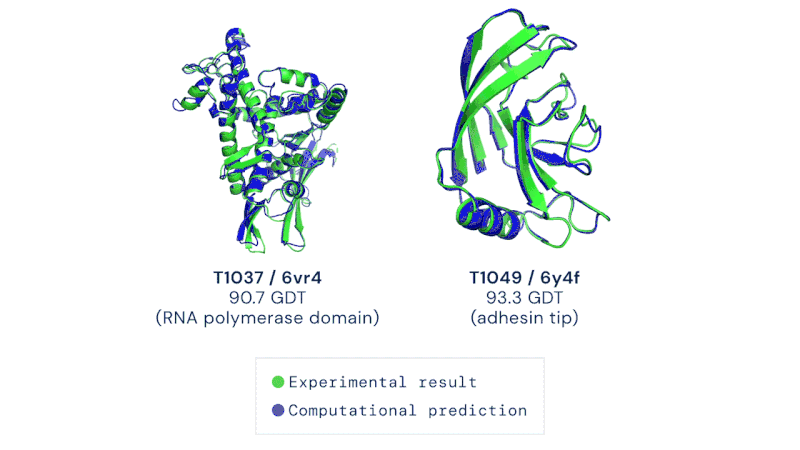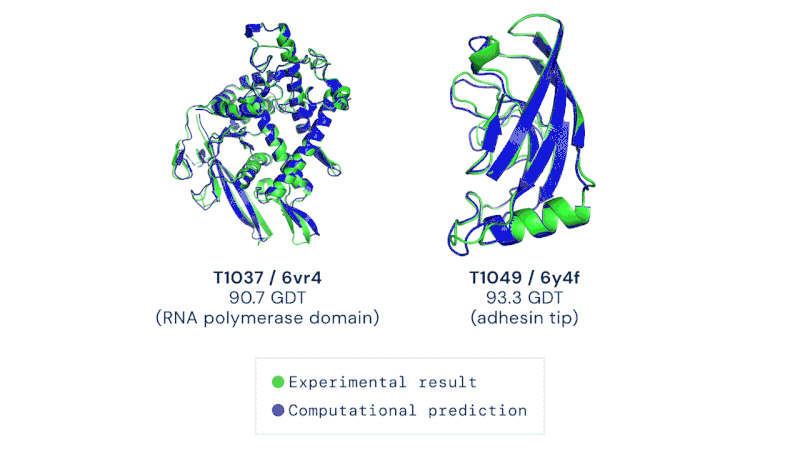
AlphaFold is an artificial intelligence (AI) program that uses deep learning to predict the 3D structure of proteins. Developed by DeepMind, a London-based subsidiary of Google, it made headlines in November 2020 when competing in the Critical Assessment of Structure Prediction (CASP). This worldwide challenge is held every two years by the scientific community and is the most well-known protein modelling benchmark. Participants must "blindly" predict the 3D structures of different proteins, and their computational methods are subsequently compared with real-world laboratory results. The CASP challenge has been held since 1994 and uses a metric known as the Global Distance Test (GDT), ranging from 0 to 100. Winners in previous years had tended to hover around the 30 to 40 mark, with a score of 90 considered to be equivalent to an experimentally determined result. In 2018, however, the team at DeepMind achieved a median of 58.9 for the GDT and an overall score of 68.5 across all targets, by far the highest of any algorithm. Then in 2020, version 2.0 of their AlphaFold program competed in the CASP, winning once again – this time with even greater accuracy. The AlphaFold 2.0 achieved a median of 92.4 across all targets, with its average margin of error comparable to the width of an atom (0.16 nanometres). Andrei Lupas, biologist at the Max Planck Institute in Germany who assessed the performances of each team in CASP, said of AlphaFold: "This will change medicine. It will change research. It will change bioengineering. It will change everything."
This month, DeepMind has announced further progress. Its latest version of AlphaFold is now 16 times faster than last year. More importantly, the company has used its AI to generate 3D structures for 350,000 proteins and made this data freely available on a new database. Until now, only 180,000 protein structures existed in the public domain, so the DeepMind computations have effectively doubled that number. The new data covers 20 different organisms – including animals such as mice, fruit flies, and bacteria like E. coli. Importantly, it also includes 98.5% of the 20,000 or so proteins in the human body. "We believe it's the most complete and accurate picture of the human proteome to date," said Dr Demis Hassabis, CEO and co-founder. "We believe this work represents the most significant contribution AI has made to advancing the state of scientific knowledge to date. And I think it's a great illustration and example of the kind of benefits AI can bring to society. We're just so excited to see what the community is going to do with this." Proteins are – to quote the famous Q from Star Trek: TNG – the building blocks of what we call life. Their formation begins with amino acids, which combine into peptides and longer polypeptides, then fold themselves into proteins. These structures are essential to biological processes. To take just one example: haemoglobin is a protein in red blood cells that carries oxygen to your body's organs and tissues and transports carbon dioxide from your organs and tissues back to your lungs. For decades, the complexity of protein folding has proven to be an immense challenge, extremely time-consuming and expensive for researchers. The AI developed by DeepMind is arguably the most significant breakthrough in the field, allowing six months of lab work to be completed in minutes. The new database – totalling about 50 gigabytes in size – will enable scientists around the world to gain faster and more accurate insights into these molecules, their 3D structures and interactions. After this first release, DeepMind plans to keep adding to the data, with a goal of releasing 100 million protein structures by the end of this year. A paper on the company's latest work appears this month in Nature. "Protein structures can provide invaluable information, both for reasoning about biological processes and for enabling interventions such as structure-based drug development or targeted mutagenesis," the authors write. "After decades of effort, 17% of the total residues in human protein sequences are covered by an experimentally-determined structure. Here we dramatically expand structural coverage by applying the state-of-the-art machine learning method, AlphaFold2, at scale to almost the entire human proteome. [...] We anticipate that routine large-scale and high-accuracy structure prediction will become an important tool, allowing new questions to be addressed from a structural perspective." "This is a Rosetta Stone moment for biology," said David Friedberg, CEO of The Production Board, a U.S. company that invests in cutting-edge health, biotech and life sciences projects. "DeepMind has translated the human genetic code into the physical machines (proteins) that run our body. Hundreds of new therapeutics will emerge as scientists map how molecules may interact with proteins to alter human health." The applications of AlphaFold go beyond medicine, however. It could help in the synthesis of novel enzymes that break down waste materials, for example, or in the production of crops that are resistant to extreme weather. "I think we're at a really exciting moment," said Dr Hassabis. "In the next decade, we, and others in the AI field, are hoping to produce amazing breakthroughs that will genuinely accelerate solutions to the really big problems we have here on Earth."
Comments »
If you enjoyed this article, please consider sharing it:
|