17th September 2019 CRISPR removes antibiotic resistance gene from bacterium Using CRISPR, researchers in the U.S. have engineered a plasmid to remove an antibiotic resistance gene from the Enterococcus faecalis bacterium. This breakthrough could lead to new methods for fighting antibiotic resistance – a threat which, on current trends, has the potential to surpass cancer in terms of mortality rate, with more than 10 million deaths each year by 2050. The research appears in Antimicrobial Agents and Chemotherapy, a journal of the American Society for Microbiology. In vitro, and in mouse models, the engineered plasmid removed the antibiotic resistance gene from E. faecalis. This microorganism is now resistant to many commonly-used antibiotics and causes life-threatening infections, especially in hospital environments. In mouse models, the plasmid reduced the abundance of the resistance gene threefold. "Our concern with organisms that cause hospital-acquired infections that are resistant to many clinical antibiotic therapies motivated the research," said senior author Breck Duerkop, PhD, Assistant Professor of Immunology and Microbiology at the University of Colorado School of Medicine.
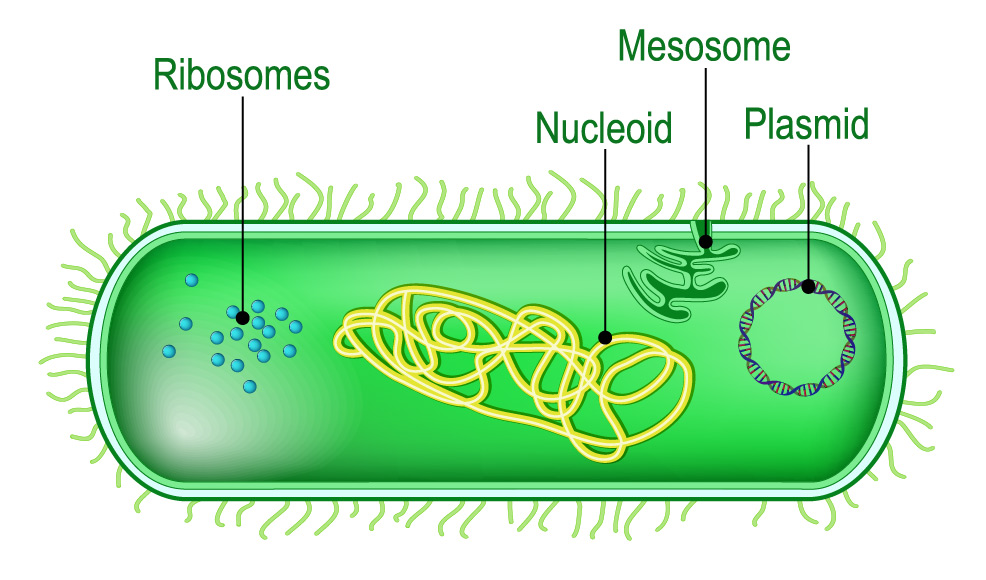
E. faecalis is part of the normal, benign intestinal flora, but when antibiotics kill off beneficial intestinal flora, E. faecalis can become pathogenic. As such, it can also acquire single or multidrug resistance. Antibiotic-resistant E. faecalis infections are today a major problem in hospitals. The mechanism used to delete antibiotic resistance genes is the specialised protein, CRISPR-Cas9, which is able to make cuts just about anywhere in DNA. Alongside CRISPR-Cas9, RNA sequences homologous to DNA within the antibiotic resistance gene have been added to the engineered plasmid. These RNAs guide the CRISPR-Cas9 to make the cuts in the right places. Plasmids are small DNA molecules within a cell that are physically separated from chromosomal DNA and can replicate independently. They are most commonly found as circular, double-stranded DNA molecules in bacteria, as shown in the illustration below.
The delivery vehicle for the engineered plasmid is a particular strain of E. faecalis, which conjugates with E. faecalis of various different strains. Conjugation is the process whereby bacteria come together to transfer genetic material from one to the other, via direct cell-to-cell contact. "E. faecalis strains used to deliver these plasmids to drug-resistant strains are immune to acquiring drug-resistant traits carried by the target cells," explains Duerkop. "The engineered plasmid can significantly reduce the occurrence of antibiotic resistance in the target bacterial population, rendering it more susceptible to antibiotics. We envision that this type of system could be used to re-sensitise antibiotic-resistant E. faecalis to antibiotics," he said. Nonetheless, Dr. Duerkop cautions that it remains possible that E. faecalis could still circumvent the engineered plasmid. Some bacteria have anti-CRISPR systems that can block CRISPR-Cas9 function, and some others have systems that can degrade foreign DNA. "Future studies will need to be done to address such an issue as E. faecalis avoiding the targeting system and under what conditions this may happen," said Dr. Duerkop.
Comments »
If you enjoyed this article, please consider sharing it:
|