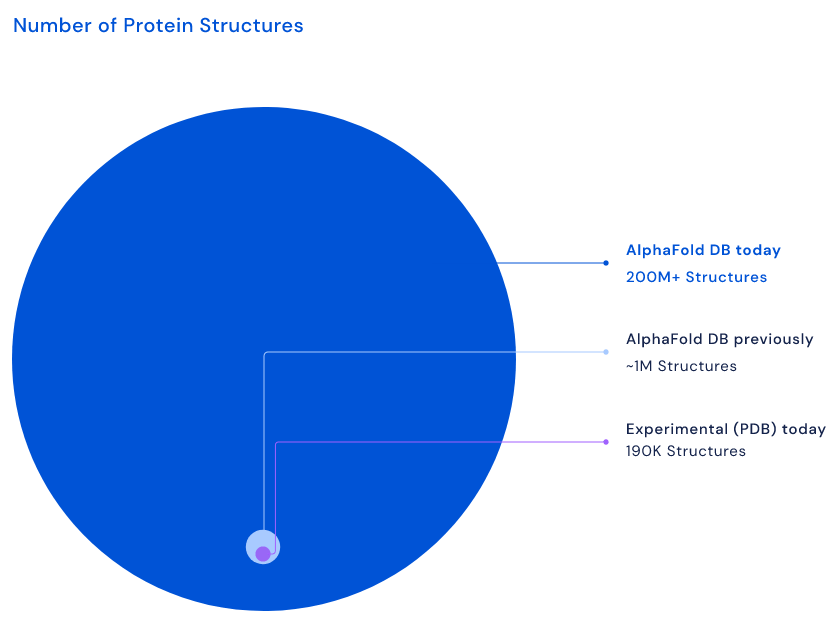
29th July 2022 AI visualises every protein known to science DeepMind's AlphaFold program has uncovered the structures of more than 200 million proteins, essentially all of those known to science.
London-based company DeepMind has reported a major milestone in biology. Scientists used one of its AI programs, AlphaFold, to accurately predict the structure of almost every protein known to science. Proteins are the building blocks of life. They begin as amino acids, which combine into peptides and polypeptides, then fold themselves into proteins. They are essential to biological processes. To take just one example: haemoglobin is a protein in red blood cells that carries oxygen to your body's organs and tissues and transports carbon dioxide from your organs and tissues back to your lungs. Version 1 of AlphaFold emerged in 2018, when it achieved first place in the Critical Assessment of protein Structure Prediction (CASP), a competition run by the global scientific community every two years. Participants in CASP must "blindly" predict the 3D structures of different proteins, and their computational methods are subsequently compared with real-world laboratory results. Across all targets, AlphaFold scored 68.5 (out of a possible 100). Version 2 won in 2020, this time with a much greater accuracy of 92.4 and a margin of error comparable to the width of an atom (0.16 nanometres). The result made headlines, as it meant that the 50-year "protein folding problem" – one of the biggest mysteries in biology – had finally been solved. DeepMind launched an even more powerful version in 2021 with 16 times the calculating speed. It generated 3D structures for a million proteins, which the company made freely available on a new database. Until then, only 180,000 protein structures had existed in the public domain, so the new data increased that number by 5.6 times. AlphaFold itself then became open-sourced, allowing scientists around the world to benefit from its abilities.
Now, the AI has taken its achievements to a whole new level. Less than 12 months after its public release, AlphaFold has been accessed by more than half a million researchers, who together have generated structures for nearly all catalogued proteins known to science: 200 million of them. "Essentially, you can think of it as covering the entire protein universe," said Demis Hassabis, co-founder and CEO of DeepMind. "It includes predictive structures for plants, bacteria, animals, and many other organisms, opening up huge new opportunities for AlphaFold to have an impact on important issues, such as sustainability, food insecurity, and neglected diseases." "AlphaFold protein structure predictions are already being used in a myriad of ways," said Professor Dame Janet Thornton, group leader at the European Bioinformatics Institute and one of the world's leading researchers in structural bioinformatics. "I expect that this latest update will trigger an avalanche of new and exciting discoveries in the months and years ahead – and this is all thanks to the fact that the data are available openly for all to use." "AlphaFold is the singular and momentous advance in life science that demonstrates the power of AI. Determining the 3D structure of a protein used to take many months or years. It now takes seconds," said Eric Topol, Founder and Director of the Scripps Research Translational Institute. "AlphaFold has already accelerated and enabled massive discoveries, including cracking the structure of the nuclear pore complex. And with this new addition of structures illuminating nearly the entire protein universe, we can expect more biological mysteries to be solved each day."
Comments »
If you enjoyed this article, please consider sharing it:
|